RC-PCR CLASSIFIER¶
Version: V0.2 (BETA)¶
install¶
-
obtain docker image
docker pull jonovox/nextflowcentos:latest -
download this project
-
extract this project
-
navigate to project
-
Download prebuild conda.tar.gz https://surfdrive.surf.nl/files/index.php/s/5q2feFVult4v81k
-
Extract conda.tar.gz in folder
single sample docker¶
sh docker/run.sh RC jonovox/nextflowcentos:latest
batch run docker¶
# USAGE
cd project
# USAGE
# bash run_batch_docker.sh <inputpath> <file_extension> <database> <threads> <image> <outputname>
# bash run_batch_docker.sh ${1} ${2} ${3} ${4} ${5} ${6}
# Example:
bash run_batch_docker.sh /workflow/input/ _001.fastq.gz SILVA 8 jonovox/easyseq_covid19:latest SILVA_test
# <file_extension> most common _001.fastq.gz
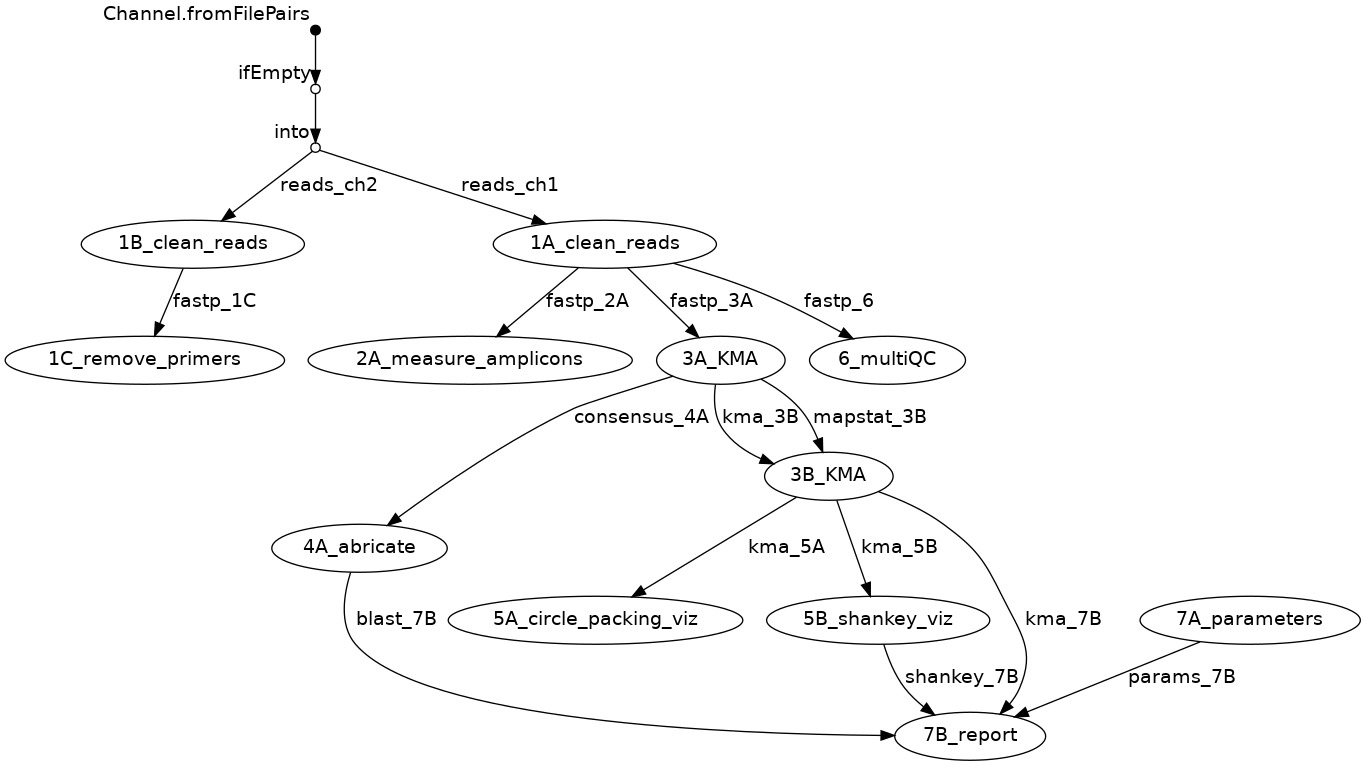
FLOW-DIAGRAM¶
conda environments¶
- 1A_clean_reads (fastp, version 0.20.1)\ env-f07c78eef9e8319c7eb087d931e36003
- 2A_measure_amplicons\ env-cd4ea0676bf53b5d7e5c6c6c523f0013
- 3A_KMA (version 1.3.28)\ env-84e06c5335c0a958ed012db619fdfceb
- 3B_process_KMA\ env-4ca5b26b8a059c60e73996439311c22f
- 4A_abricate\ env-b415f051979c22cdef40a3cbee1f0aa3
- 5A_annotation\ env-9f6b61e20675ae28786fdb538092d4db
- 6_multiQC (version 1.12)\ env-3abca7a24ea4d6c708bf4c6cea6413d2
output¶
SHANKEY PLOT¶

.
├── QC
│ └── multiqc_report.html
├── abricate
│ └── test_blast.txt #abricate/blast result
├── annotation
│ └── test.final.vcf #annotated vcf file
├── fastp
│ └── test.fastp.json
├── kma
│ ├── test.aln
│ ├── test.frag.gz
│ ├── test.frag_raw.gz
│ ├── test.fsa
│ ├── test.mapstat
│ ├── test.res #KMA result file
│ ├── test.sam
│ ├── test.sorted.bam #bam for genomebrowser
│ ├── test.sorted.bam.bai
│ └── test.vcf.gz
└── test_UMI_counttable.xlsx #primer count table
database structure¶
db
├── databasename
│ ├── KMA
│ │ ├── databasename.comp.b
│ │ ├── databasename.length.b
│ │ ├── databasename.name
│ │ └── databasename.seq.b
│ ├── blast
│ │ ├── sequences.fasta
│ │ ├── sequences.fasta.fai
│ │ ├── sequences.nhr
│ │ ├── sequences.nin
│ │ └── sequences.nsq
│ └── primers
│ └── databasename_primers.fasta
Extra¶
blastdb¶
18S
makeblastdb -in /workflow/db/blast_db/18S/sequences.fasta -title 18S -dbtype nucl -out /workflow/db/blast_db/18S/sequences
CYP51A (Afu4g06890)
makeblastdb -in /workflow/db/blast_db/CYP51A/sequences.fasta -title CYP51A -dbtype nucl -out /workflow/db/blast_db/CYP51A/sequences
CYP51A¶
18S
kma_index -i /workflow/db/KMA/18S.fa -o /workflow/db/KMA/18S
kma_index -i /workflow/db/KMA/CYP51A.fa -o /workflow/db/KMA/CYP51A
snpEff¶
manual CYP51A (Afu4g06890)
snpEff build -gff3 CYP51A
SILVA database¶
SILVA_138.1_SSURef_NR99_tax_silva_trunc convert rRNA to DNA
perl -pe 'tr/tU/uT/ unless(/>/)' < db/SILVA_138.1_SSURef_NR99_tax_silva_trunc.fasta > SILVA_138.1_SSURef_NR99_tax_silva_trunc_DNA.fasta
NOTES¶
splitting of samples on UMI using seqkit seqkit grep -irp UMI samplename.fastq.gz > output.fastq